binfalse
First SUN Spot results
May 17th, 2010One week passed since I got a package of Spots, this weekend I found some time to hack a little bit with this funny components.
First of all I programmed a tool that visualizes the Spots movement in an OpenGL frame that draws a virtual Spot. Nice for demonstrations, but nothing spectacular.
After that I developed a little mouse emulator, that translates Spot movement to mouse motions on the screen. Here the Spot isn’t doing anything intelligent, it only sends its tilt status every 25 ms as well as switch events to broadcast. Another Spot, working as basestation connected to my machine, is listening to this talking Spot and my host analyzes the received values. To move the mouse on the screen or to generate a click I use the Robot class of the Java AWT package. Long story short, a video may explain it more understandable (via YouTube):
I will continue with working on these libraries before I publish them in another post. So look forward to the release ;-)
Java network connection on Debian:SID
May 15th, 2010The unstable release of Debian is of course tricky in a lot of cases, so there is also a little stumbling stone on your path of Java network programming. On every new system it annoys me.
Before I wrongful blame my preferred Debian release called Sid I have to acknowledge I don’t know whether this feature is also available in other releases… Here is a small program to test/reproduce:
import java.net.URL;
import java.io.BufferedReader;
import java.io.InputStreamReader;
public class WebReader
{
public static void main (String[] args)
throws Exception
{
BufferedReader reader = new BufferedReader(
new InputStreamReader (new URL (args[0]).openStream ()));
String line = reader.readLine ();
while ((line = reader.readLine ()) != null)
System.out.println (line);
}
}Compilation shouldn’t fail, but if you try to launch it you’ll get an exception like that:
Exception in thread "main" java.net.SocketException: Network is unreachable
at java.net.PlainSocketImpl.socketConnect(Native Method)
at java.net.PlainSocketImpl.doConnect(PlainSocketImpl.java:333)
at java.net.PlainSocketImpl.connectToAddress(PlainSocketImpl.java:195)
at java.net.PlainSocketImpl.connect(PlainSocketImpl.java:182)
at java.net.SocksSocketImpl.connect(SocksSocketImpl.java:366)
at java.net.Socket.connect(Socket.java:525)
at java.net.Socket.connect(Socket.java:475)
at sun.net.NetworkClient.doConnect(NetworkClient.java:163)
at sun.net.www.http.HttpClient.openServer(HttpClient.java:394)
at sun.net.www.http.HttpClient.openServer(HttpClient.java:529)
at sun.net.www.http.HttpClient.<init>(HttpClient.java:233)
at sun.net.www.http.HttpClient.New(HttpClient.java:306)
at sun.net.www.http.HttpClient.New(HttpClient.java:323)
at sun.net.www.protocol.http.HttpURLConnection.getNewHttpClient(HttpURLConnection.java:860)
at sun.net.www.protocol.http.HttpURLConnection.plainConnect(HttpURLConnection.java:801)
at sun.net.www.protocol.http.HttpURLConnection.connect(HttpURLConnection.java:726)
at sun.net.www.protocol.http.HttpURLConnection.getInputStream(HttpURLConnection.java:1049)
at java.net.URL.openStream(URL.java:1010)
at WebReader.main(WebReader.java:10)This is caused by one little line in /etc/sysctl.d/bindv6only.conf saying you want to explicitly bind via IPv6. But my connection (maybe yours too) communicates still over IPv4, so this method of networking of course fails. To change this behavior you have to choose between two solutions.
Solution 1: Permanent modification (needs to be root)
You can change this behavior for the whole system by editing the file /etc/sysctl.d/bindv6only.conf :
# original only IPv6
# net.ipv6.bindv6only = 1
net.ipv6.bindv6only = 0After that just type invoke-rc.d procps restart in your terminal to let your changes take effect. Your next run should work fine.
Solution 2: Change it for this single example
If your are not allowed to change system settings, you can add -Djava.net.preferIPv4Stack=true to your execution command:
java -Djava.net.preferIPv4Stack=true WebReader http://localhost
# instead of `java WebReader http://localhost`This causes your actual runtime to connect the network via IPv4, no matter to system preferences. I hope this could save some time of developers like me ;-)
You don't know the flash-trick?
May 14th, 2010Just sitting around with Micha on a SunRay (maybe meanwhile OracleRay?). He is surfing through the web until his session seems to hang and he said:
Fuck FLASH!! Need the flash-trick...
I didn’t heard about that trick before, but now he told me that feature.
If Flash kills your SunRay session you have to type Ctrl+Alt+Moon , relogin and your session will revive. With running Flash!
As far as I know this happens very often when he is using his browser because unfortunately the whole web is contaminated with this fucking Flash… The Flash-Trick is very nice, but a flashblock plugin would be more user friendly!?
Playing around with SUN Spots
May 9th, 2010My boss wants to present some cool things in a lecture that can be done with SUN Spots. I'm selected to program these things and now I have three of them to play a little bit.
The installation was basically very easy, all you should know is that there is no chance for 64bit hosts and also Virtual Box guests don't work as expected, virtual machines lose the connection to the Spot very often... So I had to install a 32bit architecture on my host machine (btw. my decision was a Sidux Μόρος).
If a valid system is found, the rest is simple. Just download the SPOTManager from sunspotworld.com, that helps you installing the Sun SPOT Software Development Kit (SDK). If it is done connect a Sport via USB, open the SPOTManager and upgrade the Spot's software (it has to be the same version as installed on your host). All important management tasks can be done with this tool and it is possible to create virtual Spots.
Additionally to the SDK you'll get some demos installed, interesting and helpful to see how things work. In these directories ant is configured to do that crazy things that can be done with the managing tool. Here are some key targets:
ant info # get some info about the spot (version, installed application and so on)
ant deploy # build and install to spot
ant host-run # build a host application and launch it
ant help # show info about existing targets
# to configure a spot to run as base station OTA has to be disabled and basestation must be started
ant disableota startbasestationA basestation is able to administrate other Spots, so you don't have to connect each to your machine.
Ok, how to do own stuff?
There are some Netbeans plugins that makes live easier, but I don't like that big IDE's that are very slow and bring a lot of overhead to your system. To create an IDE independent project that should run on a Spot you need an environment containing:
- File: ./resources/META-INF/MANIFEST.MF
MIDlet-Name: NAME MIDlet-Version: 1.0.0 MIDlet-Vendor: Sun Microsystems Inc MIDlet-1: App Description, ,your.package.MainClassName MicroEdition-Profile: IMP-1.0 MicroEdition-Configuration: CLDC-1.1 - File: ./build.xml
< ?xml version="1.0" encoding="UTF-8" standalone="no"?> <project basedir="." default="deploy"> <property file="${user.home}/.sunspot.properties"/> <import file="${sunspot.home}/build.xml"/> </project> - Directory: ./src
Here you can place your source files
And now you can just type `ant` and the project will be deployed to the Spot.
A project that should run on your host communicating with other spots through the basestation needs a different environment:
- File: ./build.xml
< ?xml version="1.0" encoding="UTF-8" standalone="no"?> <project basedir="." default="host-run"> <property name="main.class" value="your.package.MainClassName"/> <property file="${user.home}/.sunspot.properties"/> <import file="${sunspot.home}/build.xml"/> </project> - Directory: ./src
Here you can place your source files
Ok, that's it for the moment. I'll report results.
April fools month
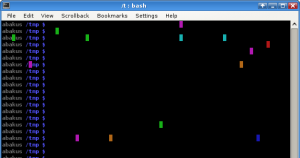
May 3rd, 2010About one month ago, it was April 1st, I attached two more lines to the .bashrc of Rumpel (he is co-worker and has to operate that day).
These two lines you can see here:
# kleiner april-scherz von dein freund martin :P
export PROMPT_COMMAND='if [ $RANDOM -le 32000 ]; then printf "\\0337\\033[%d;%dH\\033[4%dm \\033[m\\0338" $((RANDOM%LINES+1)) $((RANDOM%COLUMNS+1)) $((RANDOM%8)); fi'With each appearance of the bash prompt this command paints one pixel in the console with a random color. No respect to important content beyond this painting. That can really be annoying and he was always wondering why this happens! For more than one month, until now!
Today I lift the secret, so Rumpel, I’m very sorry ;)