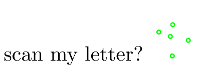binfalse
Clean Your Debian
June 14th, 2016Every once in a while you’re trying new software - maybe because you’re touching a new field, like video editing, or just to give alternatives (browsers, text editors, …) a try. However, after some time you feel your system gets messed up and the root partition, especially on notebooks, fills up with tools you don’t need anymore. To free some space you may want to get rid of all the unnecessary tools.
Clean Cached Packages
In a first step you may clean previously downloaded and cached packages.
Every time you install or upgrade something apt-getand aptitude download the package to a cache directory (e.g. /var/cache/apt/archives).
Those packages will remain there until you delete them manually - and if you upgrade your \(\LaTeX\) installation a few times this directory will grow extensively.
You can clean the cache by calling:
aptitude cleanRemove Orphaned Packages
Packages often have dependencies.
Thus, if you install a package you usually need to install other packages as well.
For example, to install gajim you also need dnsutils, python etc. (see also packages.debian.org on gajim).
If you then remove gajim, the dependencies may remain on your system.
Especially if you’re working with apt-get you may end up with orphaned packages, as apt-get remove gajim will not remove gajim’s dependencies.
To remove dependencies that are no longer required just call:
apt-get autoremove That will also help you getting rid of old kernels etc.
Find Big Packages Manually
In addition, you probably want to evaluate installed packages - to remove stuff that you do not need anymore. And you may want to remove the biggest packages first for the effort/effectiveness ratio…
In that case the following one-liner will come in handy:
dpkg-query -Wf '${Installed-Size}\t${Package}\n' | sort -nIt queries the package management system for all installed packages and requests an output format (-f) listing the package’s size and name tab-delimited per line.
Piped through sort -n the above will show you all installed packages ordered from small to huge.
Just go through the list, starting from the bottom, and delete the stuff that you do not need anymore.
That’s typically some \(\LaTeX\) stuff and old kernels and Google software etc.. ;-)
Auto-Update Debian based systems
June 5th, 2016Updating your OS is obviously super-important. But it’s also quite annoying and tedious, especially if you’re in charge of a number of systems. In about 99% it’s a monkey’s job, as it just involves variations of
aptitude update
aptitude upgradeUsually, nothing interesting happens, you just need to wait for the command to finish.
The Problem
The potential consequences in the 1% cases lets us usually swallow the bitter pill and play the monkey. The Problem is, that in some cases there is a an update that involves a modification of some configuration file that contains some adjustments of you. Let’s say you configured a daemon to listen at a specific port of your server, but in the new version they changed the syntax of the config file. That can hardly be automatised. Leaving the old version of the config will break the software, deploying the new version will dispose your settings. Thus, human interaction is required…
At least I do not dare to think about a solution on how to automatise that. But we could …
Detect the 1% and Automatise the 99%
What do we need do to prevent the configuration conflict? We need to find out which software will be updated and see if we modified one of the configuration files:
Update the package list
Updating your systems package list can be considered safe:
aptitude updateThe command downloads a list of available packages from the repositories and compares it with the list of packages installed on your system. Based on that, your update-systems knows which packages can be upgraded.
Find out which software will be updated.
The list of upgradeable packages can be obtained by doing a dry-run. The --simulate flag shows us what will be done without touching the system, -y answers every question with yes without human interaction, and -v gives us a parsable list. For example, from a random system:
root@srv » aptitude --simulate -y -v safe-upgrade
The following packages will be upgraded:
ndiff nmap
2 packages upgraded, 0 newly installed, 0 to remove and 0 not upgraded.
Need to get 4,230 kB of archives. After unpacking 256 kB will be used.
Inst nmap [6.47-3+b1] (6.47-3+deb8u2 Debian:8.5/stable [amd64])
Inst ndiff [6.47-3] (6.47-3+deb8u2 Debian:8.5/stable [all])
Conf nmap (6.47-3+deb8u2 Debian:8.5/stable [amd64])
Conf ndiff (6.47-3+deb8u2 Debian:8.5/stable [all])That tells us, the new versions of the tools nmap and ndiff will be installed. Capturing that is simple, we basically just need to grep for '^Inst'.
Check if we modified corresponding configuration files
To get the configuration files of a specific package we can ask the dpkg subsystem, for example for a dhcp client:
dpkg-query --showformat='${Conffiles}\n' --show isc-dhcp-client
/etc/dhcp/debug 521717b5f9e08db15893d3d062c59aeb
/etc/dhcp/dhclient-exit-hooks.d/rfc3442-classless-routes 95e21c32fa7f603db75f1dc33db53cf5
/etc/dhcp/dhclient.conf 649563ef7a61912664a400a5263958a6Every non-empty line contains a configuration file’s name and the corresponding md5 sum of the contents as delivered by the repository. That means, we just need to md5sum all the files on our system and compare the hashes to see if we modified the file:
# for every non-empty line
dpkg-query --showformat='${Conffiles}\n' --show $pkg | grep -v "^$" |
# do the following
while read -r conffile
do
file=$(echo $conffile | awk '{print $1}')
# the hash of the original content
exphash=$(echo $conffile | awk '{print $2}')
# the hash of the content on our system
seenhash=$(md5sum $file | awk '{print $1}')
# do thy match?
if [ "$exphash" != "$seenhash" ]
then
# STOP THE UPGRADE
# AND NOTIFY THE ADMIN
# TO MANUALLY UPGRADE THE OS
fi
doneNow we should have everything we need to compile it into a script that we can give to cron :)
The safe-upgrade script
I developed a tiny tool that can be downloaded from GitHub.. It consists of two files:
/etc/cron.daily/safeupdatescript.shis the actual acript that does the update and safe-upgrade of the system./etc/default/deb-safeupgradecan be used to overwrite the settings (hostname, mail address of the admin, etc) for a system. If it exists, the other script willsourceit.
In addition, there is a Debian package available from my apt-repository. Just install it with:
aptitude install bf-safeupgradeand let me know if there are any issues.
Disclaimer
The mentioned figure 99% is just a guess and may vary. It strongly depends on your operating system, version, and the software installed ;-)
References
mvn: Automagically create a Docker image
May 31st, 2016Having a Docker image of your software projects may make things easier for you, but will for sure lower the barrier for users to use your tools — at least in many cases ;-)
I am developing many tools in Java using Maven to manage dependencies. Thus, I’ve been looking for means to generate corresponding Docker files using the very same build management. There are already a few approaches to build Docker images through Maven, e.g. alexec’s docker-maven-plugin, and fabric8io’s docker-maven-plugin … and so on — just to name a few. However, all theses solutions seem super-heavy and they require learning new syntax and stuff, while it is so easy and doesn’t require any third party plugins.
Build Docker images using maven-antrun
Maven’s antrun plugin allows for execution of external commands. That means, you just need to maintain a proper Dockerfile along with your sources and after building the tool with maven you can call the docker-build command to create a Docker image of the current version of your tool.
I did that for a Java web application. The Dockerfile is stored as a resource in src/main/docker/Dockerfile is actually very simple:
FROM tomcat:8-jre8
MAINTAINER martin scharm
# remove the default tomcat application
RUN rm -rf /usr/local/tomcat/webapps/ROOT /usr/local/tomcat/webapps/ROOT.war
# add the BiVeS-WebApp as the new default web app
COPY BiVeS-WS-${project.version}.war /usr/local/tomcat/webapps/ROOT.warUsing Maven we can make sure (i) that the Dockerfile is copied next to the compiled and packaged tool in the target directory, (ii) that the placeholder ${project.version} in the Dockerfile is replaced with the current version of your tool, and (iii) that the docker-build command is invoked.
Copy the Dockerfile to the right place
Maven’s resources-plugin is ideally suited to deal with resources. To copy all Docker related resources to the target directory you can use the following snippet:
<plugin>
<artifactId>maven-resources-plugin</artifactId>
<executions>
<execution>
<id>copy-resources</id>
<phase>validate</phase>
<goals>
<goal>copy-resources</goal>
</goals>
<configuration>
<outputDirectory>${basedir}/target</outputDirectory>
<resources>
<resource>
<directory>src/main/docker</directory>
<filtering>true</filtering>
</resource>
</resources>
</configuration>
</execution>
</executions>
</plugin>In addition, the <filtering>true</filtering> part also makes sure to replace all Maven-related placeholders, just like the ${project.version} that we’ve been using. Thus, this solves (i) and (ii) and after the validate phase we’ll have a proper target/Dockerfile.
Build a Docker image
Using Maven’s antrun-plugin we can call the docker tool:
<plugin>
<groupId>org.apache.maven.plugins</groupId>
<artifactId>maven-antrun-plugin</artifactId>
<version>1.6</version>
<executions>
<execution>
<phase>deploy</phase>
<configuration>
<target>
<exec executable="docker">
<arg value="build"/>
<arg value="-t"/>
<arg value="binfalse/bives-webapp:${project.version}"/>
<arg value="target"/>
</exec>
</target>
</configuration>
<goals>
<goal>run</goal>
</goals>
</execution>
</executions>
</plugin>This executes a command like
docker build -t binfalse/bives-webapp:1.6.2 targetafter the deploy phase.
Thus, it builds a docker image tagged with the current version of your tool. The build’s context is target, so it will use the target/Dockerfile which COPYs the new version of your tool into the image.
Automatically build images using a Maven profile
I created a docker profile in Maven’s configuration file that is active per default if there is a src/main/docker/Dockerfile in your repository:
<profile>
<id>docker</id>
<activation>
<file>
<exists>src/main/docker/Dockerfile</exists>
</file>
</activation>
<build>
<plugins>
<plugin>
<artifactId>maven-resources-plugin</artifactId>
<!-- ... see above ... -->
</plugin>
<plugin>
<groupId>org.apache.maven.plugins</groupId>
<artifactId>maven-antrun-plugin</artifactId>
<!-- ... see above ... -->
</plugin>
</plugins>
</build>
</profile>Bonus: Also push the new image to the Docker Hub
To also push the image you need to execute the push command:
docker push binfalse/bives-webapp:1.6.2And due to the latest-confusion of Docker you also should create the latest-alias and also push that:
docker tag -f binfalse/bives-webapp:1.6.2 binfalse/bives-webapp:latest
docker push binfalse/bives-webapp:latestHowever, both is easy. Just append a few more exec calls in the antrun-plugin!
The final pom.xml snippet can be found on GitHub.
Supplement
The image for this article was derived from Wikipedia’s Apache Logo and Wikipedia’s Docker logo, licensed under the Apache License, Version 2.0.
Create an Unscanable Letter
February 8th, 2016Some time ago I’ve heard about the EURion constellation. Never heard about it? Has nothing to do with stars or astrology. It’s the thing on your money! :)
Take a closer look at your bills and you’ll discover plenty of EURions, as shown in the picture on the right. Just a few inconspicuous dots. So what’s it all about? The EURion constellation is a pattern to be recognized by imaging software, so that it can recognize banknotes. It was invented to prevent people from copying money :)
But I don’t know of any law that prohibits using that EURion, so I’ve been playing around with it. Took me some trials to find the optimal size, but I was able to create a \(LaTeX\) document that includes the EURion. That’s the essential tex code:
wanna scan my letter?
\includegraphics[width=7mm]{EURion.pdf}The whole \(LaTeX\) environment can be found on GitHub, together with the EURion image and stuff. I also provide the resulting letter.
Of course I immediately asked some friends to try to scan the letter, but it turns out, that not all scanners/printers are aware of the EURion… So it’s a bit disappointing, but I learned another thing. Good good. And to be honest, I do not have a good use case. Why should I prevent someone from printing my letters? Maybe photographers can use the EURion in their images. Copyright bullshit or something…
Monitoring of XOS devices
February 5th, 2016This week I developed some plugins for Nagios/Icinga to monitor network devices of the vendor Extreme Networks. All these plugins receive status information of, eg. switches, via SNMP.
The Basic: Check Mem, CPU, and Fans
Checking for available memory, for the device’s temperature, for the power supplies, and for fan states is quite straight forward. You just ask the switch for the values of a few OIDs, evaluate the answer, and tell Nagios/Icinga what to do.
The Simple Network Management Protocol (SNMP) is actually a very easy to use protocol. There is an SNMP server, such as a router or a switch, which exposes management data through the SNMP protocol. To access these data you just send an object identify (OID) to an SNMP server and receive the corresponding value. So called management information bases (MIB) can tell you what a certain OID stands for.
On the command line, for example, you could use snmpwalk to iterate over an OID subtree to, e.g., obtain information about the memory on a device:
usr@srv $ snmpwalk -v 2c -c publicCommunityString switch.address.com 1.3.6.1.4.1.1916.1.32.2.2.1
1.3.6.1.4.1.1916.1.32.2.2.1.1.1 = Gauge32: 1
1.3.6.1.4.1.1916.1.32.2.2.1.2.1 = STRING: "262144"
1.3.6.1.4.1.1916.1.32.2.2.1.3.1 = STRING: "116268"
1.3.6.1.4.1.1916.1.32.2.2.1.4.1 = STRING: "7504"
1.3.6.1.4.1.1916.1.32.2.2.1.5.1 = STRING: "138372"
The OID 1.3.6.1.4.1.1916.1.32.2.2.1 addresses the memory information table of the SNMP provider at switch.address.com.
The value at *.2.1 shows how much memory is installed, *.3.1 shows how much memory is free, *.4.1 shows how much is consumed by the system, and *.5.1 shows how much is consumed by user processes. Basic calculations tell us there are 262144/1024 = 256KB in total and 100*116268/262144 = 44.35% is free.
A bit more logic for a warning/critical switch and the plugin is done.
The Feature: Monitoring of the FDB
But I would probably not write about that basic stuff if there was not an extra feature! I implemented a script to also monitor the FDB. FDB is and abbreviation for forwarding databases: The switch maintains a forwarding database (FDB) of all MAC addresses received on all of its ports. It, for example, uses the information in this database to decide whether a frame should be forwarded or filtered. Each entry consists of
- the MAC address of the device behind the port
- the associated VLAN
- the age of the entry – depending on the configuration the entries age out of the table
- some flags – e.g. is the entry dynamic or static
- the port
The table may look like the following:
> show fdb
Mac Vlan Age Flags Port / Virtual Port List
------------------------------------------------------------------------------
01:23:45:67:89:ab worknet(0060) 0056 n m 9
01:23:42:67:89:ab mobnet(0040) 0001 n m 21
Flags : d - Dynamic, s - Static, p - Permanent, n - NetLogin, m - MAC, i - IP,
x - IPX, l - lockdown MAC, L - lockdown-timeout MAC, M- Mirror, B - Egress Blackhole,
b - Ingress Blackhole, v - MAC-Based VLAN, P - Private VLAN, T - VLAN translation,
D - drop packet, h - Hardware Aging, o - IEEE 802.1ah Backbone MAC,
S - Software Controlled Deletion, r - MSRP
As soon as the switch gets a frame on one port it learns the corresponding MAC address, port number, etc. into this table. So if a frame for this MAC address arrives it know where to send it to.
However, that content of a networking class.
All we need to know is that a switch can tell you which device which MAC address is is connected to which port.
And that’s the idea of check_extreme_fdb.pl! It compares the entries of the FDB with some expected entries in an CSV file. The CSV is supposed to contain three coloumns:
mac,port,vlan
If a MAC address in the FDB matches the MAC address in the CSV file it checks the ports and vlans. If those do not match, it will raise an error.
For the CSV: Feel free to leave port or vlan empty if you do not care about this detail.
That means, if you just want to make sure that the device with the MAC 01:23:45:67:89:ab is in vlan worknet you add an entry such as:
01:23:45:67:89:ab,,worknet
Use -e <FILE> to pass the CSV file containing expected entry to the program and call it like beckham:
perl -w check_extreme_fdb.pl -s <SWITCH> -C <COMMUNITY-STRING> -e <EXPECTED>
Here, SWITCH being the switch’s address and COMMUNITY-STRING beeing the SNMP “passphrase”. You may also want to add -w to raise a warning if one of the entries in the CSV file wasn’t found in the FDB. To create a sample CSV file that matches the current FDB you can call it with --print.
To get the script have a look at the check_extreme_fdb.pl software page.
More Extreme Stuff
In addition there are some other scripts to monitor Extreme Networks devices: