binfalse
Eventually learning how to wield PAM
July 16th, 2016PAM. The Pluggable Authentication Modules. I’m pretty sure you heard of it. It sits in its /etc/pam.d/ and does a very good job. Reliable and performant, as my guts tend to tell me.
Unless… You want something specific! In my case that always implied a lot of trial and error. Copying snippets from the internet and moving lines up and down and between different PAM config files. So far, I managed to conquer that combinatorial problem in less time than I would need to learn PAM - always with bad feeling because I don’t know what I’ve been doing with the f*cking sensible auth system…
But this time PAM drives me nuts. I want to authenticate users via the default *nix passwd as well as using an LDAP server -AND- pam_mount should mount something special for every LDAP user. The trial and error method gave me two versions of the config that works for one of the tasks, but I’m unable to find a working config for both. So… Let’s learn PAM.
The PAM
On linux systems, PAM lives in /etc/pam.d/. There are several config files for differen purposes. You may change them and they will have effect immediately – no need to restart anything.
PAM allows for what they call “stacking” of modules: Every configuration file defines a stack of modules that are relevant for the authentication for an application. Every line in a config file containes three tokens:
- the realm is the first word in a line; it is either auth, account, password or session
- he control determines what PAM should do if the module either succeeds/fails
- the module the actual module that gets invoked and optionally some module parameters
Realms
There are four different realms:
- auth: checks that the user is who he claims to be; usually password base
- account: account verification functionality; for example checking group membership, account expiration, time of day if a user only has part-time access, and whether a user account is local or remote
- password: needed for updating passwords for a given service; may involve e.g. dictionary checking
- session: stuff to setup or cleanup a service for a given user; e.g. launching system-wide init script, performing special logging, or configuring SSO
Controls
In most cases the controls consist of a single keyword that tells PAM what to do if the corresponding module either succeeds or fails. You need to understand, that this just controls the PAM library, the actual module neither knows not cares about it. The four typical keywords are:
- required: if a ‘required’ module is not successful, the operation will ultimately fail. BUT only after the modules below are done! That is because an attacker shouldn’t know which or when a module fails. So all modules will be invoked even if the first on fails, giving less information to the bad guys. Just note, that a single failure of a ‘required’ module will cause the whole thing to fail, even if everything else succeeds.
- requisite: similar to required, but the whole thing will fail immediately and the following modules won’t be invoked.
- sufficient: a successful ‘sufficient’ module is enough to satisfy the requirements in that realm. That means, the following ‘sufficient’ won’t be invoked! However, sufficient modules are only sufficient, that means (i) they may fail but the realm may still be satisfied by something else, and (ii) they may all succeed but the realm may fail because a required module failed.
- optional: optional modules are non-critical, they may succeed or fail, PAM actually doesn’t care. PAM only cares if there are exclusively optional modules in a particular stack. In that case at least one of them needs to succeed.
Modules
The last token of every line lists the path to the module that will be invoked. You may point to the module using an absolute path starting with / or a path relative from PAMs search directories. The search path depends on the system your working on, e.g. for Debian it is /lib/security/ or /lib64/security/. You may also pass arguments to the module, common arguments include for example use_first_pass which provides the password of an earlier module in the stack, so the users doesn’t need to enter their password again (e.g. useful when mounting an encrypted device that uses the same password as the user account).
There are dozens of modules available, every module supporting different options. Let me just forward you to a PAM modules overview at linux-pam.org. and an O’Reilly article on useful PAM modules.
That’s it
Yeah, reading and writing about it helped me fixing my problem. This article is probably just one within a hundred, so if it didn’t help you I’d like to send you to one of the following. Try reading them, if that doesn’t help write a blog post about it ;-)
Further Resources
- Centos documents on PAM, including writing of PAM modules
- The Linux-PAM Guides
- O’Reilly’s Introduction to PAM
- The Linux-PAM System Administrators’ Guide
Modify a linux LiveOS
July 8th, 2016By default I’m using GRML when I need a live operating system. I installed it to one of my USB pen drives and (almost) always carry it with me. GRML already has most of the essential and nice-to-have tools installed and it’s super compfortable when the shit has hit the fan!
The Problem
However, there are circumstances when you need something that’s not available on the base image. That entails a bit annoying work. For example if you need to install package XXX you need to
- run
aptitude install XXX - recognize that the package lists are super out-dated…
- call
aptitude update - recognize you’re missing the latest GPG key that was used to sign the packages
- run a
gpg --keyserver keys.gnupg.net --recv YYYfollowed by agpg --export YYY | sudo apt-key add - - run
aptitude install XXXagain to get the package
And that’s actually really annoying if you nedd XXX often. You’ll wish to have XXX in the base system! But how to get it there?
Update the ISO image
The following will be based on GRML. I assume that your USB pen drive will be recognized as /dev/sdX and the partition you (will) have created is /dev/sdX1. The following also requires syslinux to be installed:
aptitude install syslinuxInstall the original Image to your Pen Drive
To modify an image you first need the image. So go to grml.org/download/ and download the latest version of the image. At the time of writing this article it is Grml 2014.11 (Gschistigschasti). You see it’s a bit outdated, wich also explains this article ;-)
To install it on a pen drive you need a pen drive. It should have a partition with a bootable flag. Use, eg., fdisk or gparted.
This partition should have a FAT partition on it. If your not using the UI of gparted this is the way to go (assuming /dev/sdX1 is the partition you created):
mkfs.fat -F32 -v -I -n "LIVE" /dev/sdX1Then you need to install syslinux’ MBR onto the pen drive:
dd if=/usr/lib/syslinux/mbr/mbr.bin of=/dev/sdXMount both, the ISO (/tmp/grml96-full_2014.11.iso) and the pen drive (/dev/sdX1) to copy all files from the ISO onto the pen drive:
mkdir -p /mnt/mountain/{iso,usb}
mount /dev/sdX1 /mnt/mountain/usb
mount -o loop,ro -t iso9660 /tmp/grml96-full_2014.11.iso /mnt/mountain/iso
rsync -av /mnt/mountain/iso/* /mnt/mountain/usb/If you now have a look into the /mnt/mountain/usb/ directory you will see the GRML live image structure. However, you won’t be able to boot – you still need a proper bootloader.
Fortunately, the syslinux tool is able to install it to your pen drive:
syslinux /dev/sdX1The syslinux bootloader still need some configuration, but GRML alreay contains them. Just copy them from the USB’s/boot/isolinux/ into the root of the pen drive:
cp /mnt/mountain/usb/boot/isolinux/* /mnt/mountain/usbExcellent! Unmount the pendrive an try to boot from it, to make sure it’s properly booting the GRML operating system:
umount /mnt/mountain/iso/ /mnt/mountain/usb/ && syncUnderstand what happens
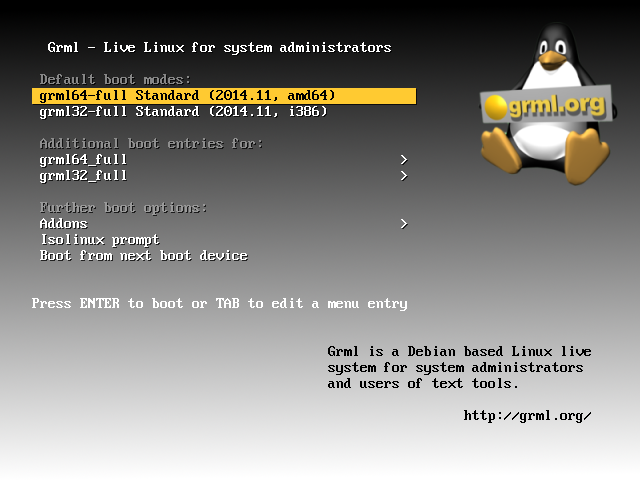
Ok, if that was successful you’re mashine will boot into the GRML splash screen (see Figure) and you can choose between the 64bit and the 32bit version (and some more specific boot options). Clicking either of them will mount a read-only file system from the pen drive as a read-write file system into your RAM. Afterwards it will boot from that sytem in the RAM.
The file system that’s written to the memory can be found in live/grml64-full/grml64-full.squashfs (or /mnt/mountain/usb/live/grml32-full/grml32-full.squashfs for 32bit systems).
It’s a squashfs, see wikipedia on SquashFS.
Modify the Base Image
So let’s go and modify the system.
You probably alreay guess it: We need to update the grml64-full.squashfs and integrate the desired changes.
As I said, the image of the file system can be found in /live/grml64-full/grml64-full.squashfs and /live/grml32-full/grml32-full.squashfs of the USB’s root.
In the following I will just use the 64bit version in /live/grml64-full/grml64-full.squashfs, but it can be applied to the 32bit version euqivalently – just update the paths.
As the SquashFS is read-only you need to mount it and copy all the data to a location on a read-write file system.
To copy the stuff preserving all attributes I recommend using the rsync tool.
Let’s assume your pen drive is again mounted to /mnt/mountain/usb and /storage/modified-livecd is mounted as RW:
mount /mnt/mountain/usb/live/grml64-full/grml64-full.squashfs /mnt/mountain/iso -t squashfs -o loop
rsync -av /mnt/mountain/iso/* /storage/modified-livecd/Now you have your system read-write in /storage/modified-livecd. Go ahead and modify it! For example, create a new file:
echo "this was not here before" > /storage/modified-livecd/NEWFILEThen you just need to delete the original SquashFS file and repack the new version onto the pen drive:
rm /mnt/mountain/usb/live/grml64-full/grml64-full.squashfs
mksquashfs /storage/modified-livecd /mnt/mountain/usb/live/grml64-full/grml64-full.squashfsVoilà… Just boot from the pen drive and find your modified live image :)
Of course you can also chroot into /storage/modified-livecd to e.g. install packages and modify it from inside…
For those not familiar with chrooting: [Wikipedia comes with a good example on how to chroot.](https://en.wikipedia.org/wiki/Chroot#Linux_host_kernel_virtual_file_systems_and_configuration_files
So… Conclusion?
Yeah, it’s not that hard to modify the system, but it takes some manual steps. And if you plan to do it more often you need to think about infrastructure etc. Thus, this is probably just applicable for hard-core users – but those will love it ;-)
PHP file transfer: Forget the @ - use `curl_file_create`
June 21st, 2016I just struggled uploading a file with PHP cURL. Basically, sending HTTP POST data is pretty easy. And to send a file you just needed to prefix it with an at sign (@). Adapted from the most cited example:
<?php
$target_url = 'http://server.tld/';
$post = array (
'extra_info' => '123456',
'file_contents' => '@' . realpath ('./sample.jpeg');
);
$ch = curl_init ($target_url);
curl_setopt ($ch, CURLOPT_POST, 1);
curl_setopt ($ch, CURLOPT_POSTFIELDS, $post);
curl_setopt ($ch, CURLOPT_RETURNTRANSFER, 1);
$result=curl_exec ($ch);
curl_close ($ch);
?>You see, if you add an ‘@’ sign as the first character of a post field the content will be interpreted as a file name and will be replaced by the file’s content.
At least, that is how it used to be… And how most of the examples out there show you.
However, they changed the behaviour. They recognised that this is obviously inconvenient, insecure and error prone. You cannot send POST data that starts with an @ and you always need to sanitise user-data before sending it, as it otherwise may send the contents of files on your server. And, thus, they changed that behaviour in version 5.6, see the RFC.
That means by default the @/some/filename.ext won’t be recognized as a file – PHP cURL will literally send the @ and the filename (@/some/filename.ext) instead of the content of that file. Took ma a while and some tcpdumping to figure that out..
Instead, they introduced a new function called curl_file_create that will create a proper CURLFile object for you. Thus, you should update the above snippet with the following:
<?php
$target_url = 'http://server.tld/';
$post = array (
'extra_info' => '123456',
'file_contents' => curl_file_create ('./sample.jpeg');
);
$ch = curl_init ($target_url);
curl_setopt ($ch, CURLOPT_POST, 1);
curl_setopt ($ch, CURLOPT_POSTFIELDS, $post);
curl_setopt ($ch, CURLOPT_RETURNTRANSFER, 1);
$result=curl_exec ($ch);
curl_close ($ch);
?>Note that the contents of the file_contents field of the $post data differs.
The php.net manual for the file_contents function is unfortunatelly not very verbose, but the RFC on wiki.php.net tells you a bit more about the new function:
<?php
/**
* Create CURLFile object
* @param string $name File name
* @param string $mimetype Mime type, optional
* @param string $postfilename Post filename, defaults to actual filename
*/
function curl_file_create($name, $mimetype = '', $postfilename = '')
{}
?>So you can also define a mime type and some file name that will be sent.
That of course makes it a bit tricky to develop for different platforms, as you want your code to be valid on both PHP 5.4 and PHP 5.6 etc. Therefore you could introduce the following as a fallback:
<?php
if (!function_exists('curl_file_create'))
{
function curl_file_create($filename, $mimetype = '', $postname = '')
{
return "@$filename;filename="
. ($postname ?: basename($filename))
. ($mimetype ? ";type=$mimetype" : '');
}
}
?>as suggested by mipa on php.net. This creates a dummy function that is compliant with the old behaviour if there is no curl_file_create, yet.
Clean Your Debian
June 14th, 2016Every once in a while you’re trying new software - maybe because you’re touching a new field, like video editing, or just to give alternatives (browsers, text editors, …) a try. However, after some time you feel your system gets messed up and the root partition, especially on notebooks, fills up with tools you don’t need anymore. To free some space you may want to get rid of all the unnecessary tools.
Clean Cached Packages
In a first step you may clean previously downloaded and cached packages.
Every time you install or upgrade something apt-getand aptitude download the package to a cache directory (e.g. /var/cache/apt/archives).
Those packages will remain there until you delete them manually - and if you upgrade your \(\LaTeX\) installation a few times this directory will grow extensively.
You can clean the cache by calling:
aptitude cleanRemove Orphaned Packages
Packages often have dependencies.
Thus, if you install a package you usually need to install other packages as well.
For example, to install gajim you also need dnsutils, python etc. (see also packages.debian.org on gajim).
If you then remove gajim, the dependencies may remain on your system.
Especially if you’re working with apt-get you may end up with orphaned packages, as apt-get remove gajim will not remove gajim’s dependencies.
To remove dependencies that are no longer required just call:
apt-get autoremove That will also help you getting rid of old kernels etc.
Find Big Packages Manually
In addition, you probably want to evaluate installed packages - to remove stuff that you do not need anymore. And you may want to remove the biggest packages first for the effort/effectiveness ratio…
In that case the following one-liner will come in handy:
dpkg-query -Wf '${Installed-Size}\t${Package}\n' | sort -nIt queries the package management system for all installed packages and requests an output format (-f) listing the package’s size and name tab-delimited per line.
Piped through sort -n the above will show you all installed packages ordered from small to huge.
Just go through the list, starting from the bottom, and delete the stuff that you do not need anymore.
That’s typically some \(\LaTeX\) stuff and old kernels and Google software etc.. ;-)
Auto-Update Debian based systems
June 5th, 2016Updating your OS is obviously super-important. But it’s also quite annoying and tedious, especially if you’re in charge of a number of systems. In about 99% it’s a monkey’s job, as it just involves variations of
aptitude update
aptitude upgradeUsually, nothing interesting happens, you just need to wait for the command to finish.
The Problem
The potential consequences in the 1% cases lets us usually swallow the bitter pill and play the monkey. The Problem is, that in some cases there is a an update that involves a modification of some configuration file that contains some adjustments of you. Let’s say you configured a daemon to listen at a specific port of your server, but in the new version they changed the syntax of the config file. That can hardly be automatised. Leaving the old version of the config will break the software, deploying the new version will dispose your settings. Thus, human interaction is required…
At least I do not dare to think about a solution on how to automatise that. But we could …
Detect the 1% and Automatise the 99%
What do we need do to prevent the configuration conflict? We need to find out which software will be updated and see if we modified one of the configuration files:
Update the package list
Updating your systems package list can be considered safe:
aptitude updateThe command downloads a list of available packages from the repositories and compares it with the list of packages installed on your system. Based on that, your update-systems knows which packages can be upgraded.
Find out which software will be updated.
The list of upgradeable packages can be obtained by doing a dry-run. The --simulate flag shows us what will be done without touching the system, -y answers every question with yes without human interaction, and -v gives us a parsable list. For example, from a random system:
root@srv » aptitude --simulate -y -v safe-upgrade
The following packages will be upgraded:
ndiff nmap
2 packages upgraded, 0 newly installed, 0 to remove and 0 not upgraded.
Need to get 4,230 kB of archives. After unpacking 256 kB will be used.
Inst nmap [6.47-3+b1] (6.47-3+deb8u2 Debian:8.5/stable [amd64])
Inst ndiff [6.47-3] (6.47-3+deb8u2 Debian:8.5/stable [all])
Conf nmap (6.47-3+deb8u2 Debian:8.5/stable [amd64])
Conf ndiff (6.47-3+deb8u2 Debian:8.5/stable [all])That tells us, the new versions of the tools nmap and ndiff will be installed. Capturing that is simple, we basically just need to grep for '^Inst'.
Check if we modified corresponding configuration files
To get the configuration files of a specific package we can ask the dpkg subsystem, for example for a dhcp client:
dpkg-query --showformat='${Conffiles}\n' --show isc-dhcp-client
/etc/dhcp/debug 521717b5f9e08db15893d3d062c59aeb
/etc/dhcp/dhclient-exit-hooks.d/rfc3442-classless-routes 95e21c32fa7f603db75f1dc33db53cf5
/etc/dhcp/dhclient.conf 649563ef7a61912664a400a5263958a6Every non-empty line contains a configuration file’s name and the corresponding md5 sum of the contents as delivered by the repository. That means, we just need to md5sum all the files on our system and compare the hashes to see if we modified the file:
# for every non-empty line
dpkg-query --showformat='${Conffiles}\n' --show $pkg | grep -v "^$" |
# do the following
while read -r conffile
do
file=$(echo $conffile | awk '{print $1}')
# the hash of the original content
exphash=$(echo $conffile | awk '{print $2}')
# the hash of the content on our system
seenhash=$(md5sum $file | awk '{print $1}')
# do thy match?
if [ "$exphash" != "$seenhash" ]
then
# STOP THE UPGRADE
# AND NOTIFY THE ADMIN
# TO MANUALLY UPGRADE THE OS
fi
doneNow we should have everything we need to compile it into a script that we can give to cron :)
The safe-upgrade script
I developed a tiny tool that can be downloaded from GitHub.. It consists of two files:
/etc/cron.daily/safeupdatescript.shis the actual acript that does the update and safe-upgrade of the system./etc/default/deb-safeupgradecan be used to overwrite the settings (hostname, mail address of the admin, etc) for a system. If it exists, the other script willsourceit.
In addition, there is a Debian package available from my apt-repository. Just install it with:
aptitude install bf-safeupgradeand let me know if there are any issues.
Disclaimer
The mentioned figure 99% is just a guess and may vary. It strongly depends on your operating system, version, and the software installed ;-)